New Discovery about How Immune Cells Recognize Lipid Nanoparticles
Scientists from the Institute of High Energy Physics, Chinese Academy of Sciences (IHEP), in collaboration with the National Center for Nanoscience and Technology (NCNST) and Peking Univserity (PKU) teams, has made significant progress in identifying cell membrane receptors for lipid nanoparticles (LNPs). The research, titled “Identification of Cell Receptors Responsible for Recognition and Binding of Lipid Nanoparticles” was published in the Journal of the American Chemical Society on February 24. The study was co-led by Dr. Baimanov Didar (Postdoctoral Researcher, IHEP) and Dr. WANG Jing (Associate Researcher, PKU), supervised by Prof. WANG Liming (IHEP) and Prof. CHEN Chunying (NCNST) as corresponding authors.
This study introduced an innovative approach to identifying nanoparticle recognition receptors, revealing the dynamic composition of the protein corona formed on LNPs in human blood and uncovering key mechanisms underlying their interactions with immune cell receptors. These findings offer essential insights for enhancing mRNA vaccine design and pave the way for more precise nanomedicine delivery.
Research was inspired by the need to understand why some mRNA vaccines work better than others and why certain nanomedicines are cleared too quickly. Researchers sought novel methods to identify how lipid nanoparticles (LNPs) are recognized by the immune system. Once inside the body, LNPs rapidly adsorb blood proteins, forming a protein corona that determines how they interact with cells, whether they reach their target, and how fast they are cleared. However, existing methods to study these interactions often disrupt the natural corona composition, making it difficult to accurately identify which cell receptors recognize and bind to LNPs.
To overcome this challenge, based on their previous work (Nat. Commun. 2022, 13, 5389), the scientists developed a novel “Fishing” technique that enables real-time tracking of how LNPs interact with plasma proteins and immune cells in their native state. This innovative biosensor-based approach allows us to capture the full dynamic process of protein corona formation and to identify the cell receptors responsible for LNP recognition directly without altering their structure (Figure 1). Compared to traditional centrifugation-based approaches, this method better preserves LNP structural integrity, provides a more accurate reflection of the soft/hard protein corona composition, and allows for in situ identification of cell receptors recognizing the LNP protein corona.
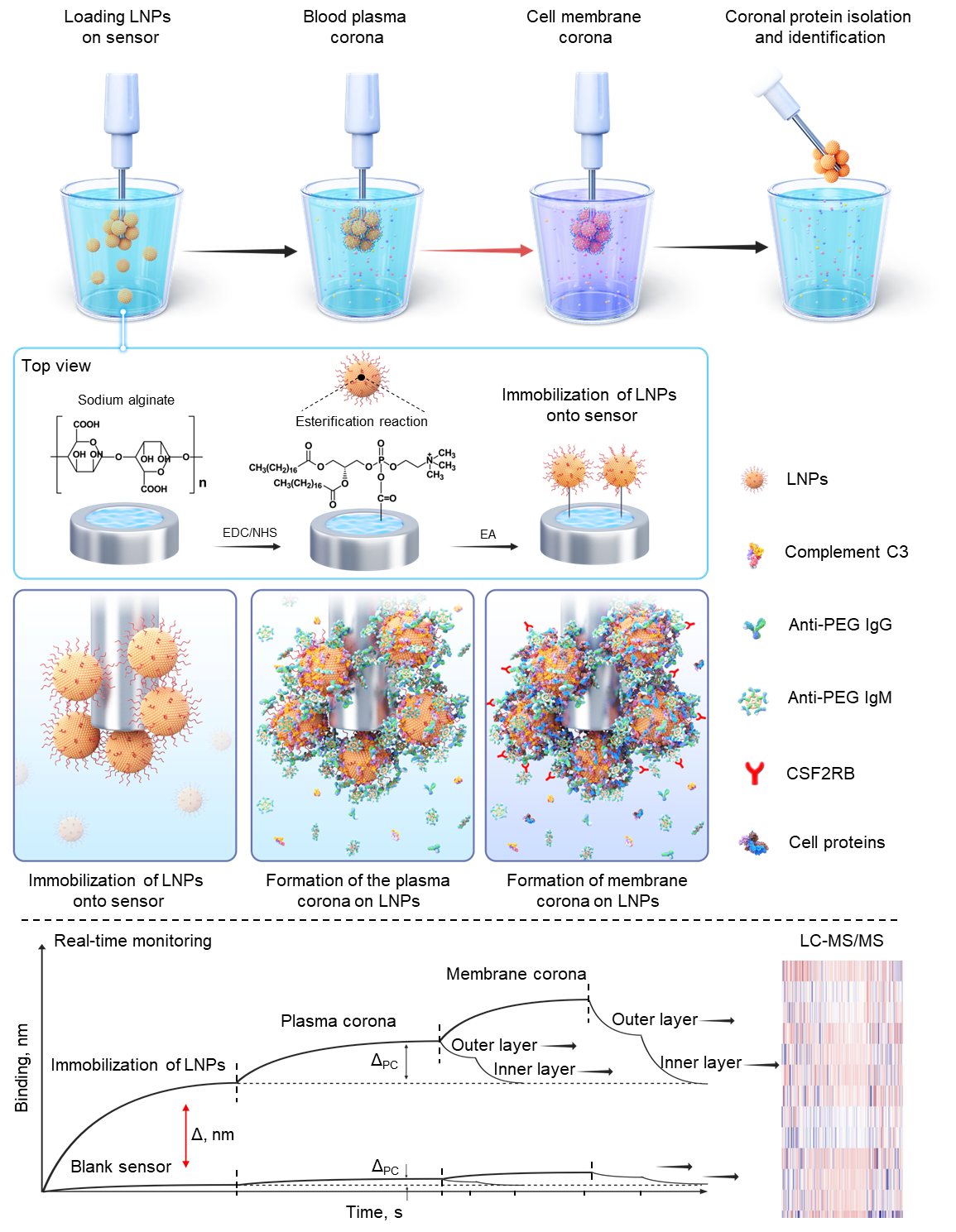
Figure 1. Development of the "Fishing" technique for identifying LNP protein corona components and cell membrane receptors.
Using this method, researchers discovered that anti-PEG antibodies, which accumulate in the body after repeated exposure to PEGylated drugs or vaccines, play a crucial role in accelerating LNP clearance. This explains why some individuals have shorter circulation times for mRNA vaccines or experience reduced drug efficacy with repeated dosing. More importantly, they identified CSF2RB (Colony-Stimulating Factor 2 Receptor Beta) as a key receptor that myeloid immune cells use to recognize and engulf LNPs (Figure 2), providing a new target for improving nanomedicine stability and delivery.
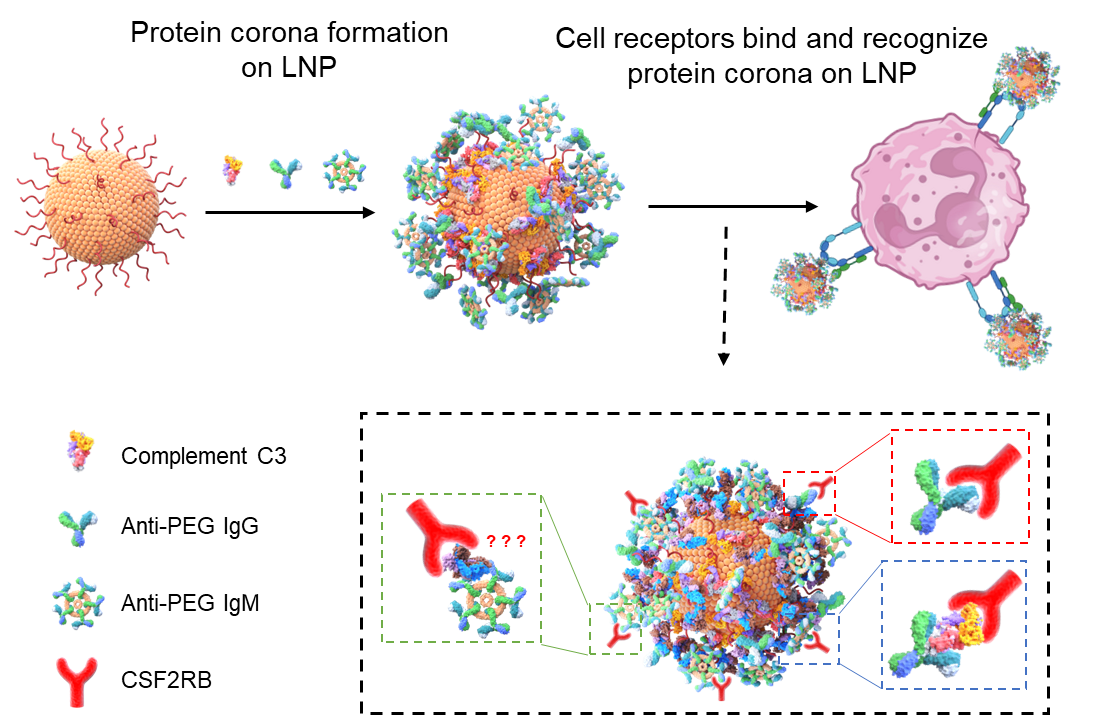
Figure 2. LNP behavior in blood driven by PEG antibodies and complement opsonins on the plasma protein corona, leading to myeloid cell receptor-mediated endocytosis.
These findings open new possibilities for engineering smarter LNP-based vaccines and therapies that are more resistant to immune clearance. Future strategies could involve designing alternative LNP coatings to replace PEG, fine-tuning immune evasion mechanisms, or targeting CSF2RB-related pathways to extend vaccine effectiveness and improve drug delivery precision.
By unraveling how nanomedicines interact with the immune system, this study lays the foundation for safer, longer-lasting, and more effective mRNA vaccines and next-generation therapies.
The research was supported by the National Key R&D Program, National Natural Science Foundation of China, and other major funding initiatives.
Paper link: https://pubs.acs.org/doi/10.1021/jacs.4c16987