| Crystallographic analysis of the conserved C-terminal domainof transcription factor Cdc73 from Saccharomyces cerevisiaereveals a GTPase-like fold |
| From: PublishDate:2013-06-15 Hits: |
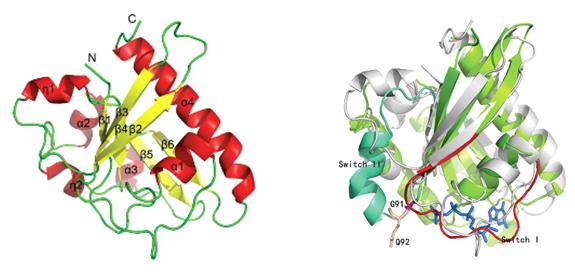
Paf1 complex which contains Paf1, Cdc73, Ctr9, Leo1 and Rtf1 (recently discovered hSki8 is one subunit in human Paf1 complex) subunits is a conserved complex in eukaryotes. The main function of Paf1 complex is taking participate into transcription including initiation, elongation and termination together with RNA polⅡ. Till now, only a little is known about the structure information of Paf1 complex. Before the structure of the C-terminal domain of Saccharomyces cerevisiae Cdc73 was reported, only the Plus-3 domain structure of human Rtf1 had been determined. The Maikun Teng and Liwen Niu groups from University of Science and Technology of China collected a set of 2.5Å diffraction data of Saccharomyces cerevisiae Cdc73 under the assistance of the staff on beamline 3W1A of the Beijing Synchrotron Radiation Facility and solved the structure by using the heavy atom iodine anomalous scattering data. The results were published in August 2012 on the journal “Acta Crystallographica Section D Biological Crystallography”. In the same period, the Karen M. Arndt, Andrew P. VanDemark groups from the University of Pittsburgh solved the segment structure of Cdc73 too. Paf1 complex as a platform on RNA polⅡthat coordinates the association of many critical transcription factors plays a key role in one or more steps in transcription. In a very recently study, the C-terminal domain of Cdc73, Rtf1 or Ctr9 were shown to directly bind to RNA polⅡ CTD repeat peptides in a phosphorylation-specific manner. As a tumor suppressor, the mutation of Human Cdc73 can lead to hyperparathyroidism-jaw tumor (HPT-JT) or other tumors. A DALI server search indicates the C-terminal structure of Cdc73 is a canonical α6/β6 GTPase-like fold, although the primary sequence of it has very low identity to GTPases and is not conservative at the key region. No significant GTP-binding ability and hydrolysis activity were detected by ITC and enzyme activity assay. So Cdc73, with a GTPase-like fold, may not be a GTPase. The determination of the structure of Cdc73 has provided further insights into the assembly way of Paf1 complex and the structural basis for study on Cdc73-RNA PolⅡ interaction and human related diseases. Further research work is underway.
Fig.1 Ribbon diagram of yCdc73_C. Fig.2 Superposition of yCdc73_C (grey) and Rab33 (green).
Article: Hongkai Chen, Nuo Shi,Yongxiang Gao, Xu Li, Maikun Teng* and Liwen Niu*, Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold. Acta Crystallogr D Biol Crystallogr. 68(Pt 8):953-9, 2012. |
|
|
| Chinese
Science Highlights
Home /
Copyright © 2011 - 2012 Beijing Synchrotron Radiation Facility